
Applications of Ligase Fidelity Data & Tools
Return to NEBridge® Golden Gate AssemblyFeatured NEB scientist publications
Learn more about NEB’s research on ligase fidelity and the use of Data-optimized Assembly Design (DAD) and Ligase Fidelity Tools in our growing list below.
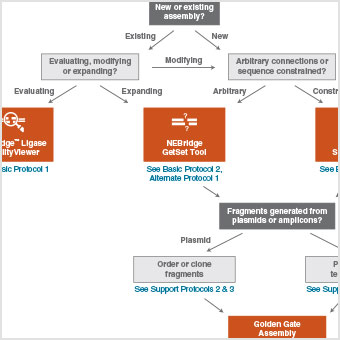 |
High-Complexity One-Pot Golden Gate Assembly
|
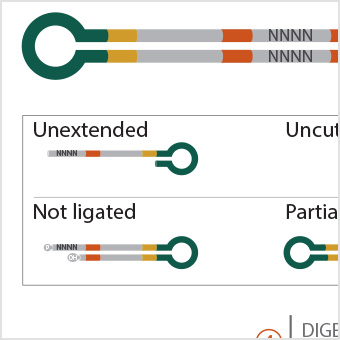 |
Profiling DNA Ligase Substrate Specificity with a Pacific Biosciences Single-Molecule Real-Time Sequencing Assay
|
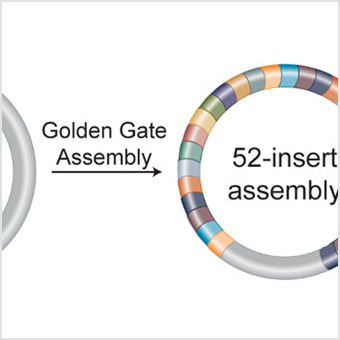 |
Rapid 40 kb Genome Construction from 52 Parts through Data-optimized Assembly Design
|
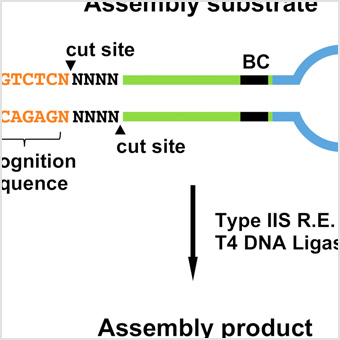 |
Enabling one-pot Golden Gate assemblies of unprecedented complexity using data-optimized assembly design
|
 |
Comprehensive Profiling of Four Base Overhang Ligation Fidelity by T4 DNA Ligase and Application to DNA Assembly
|
 |
A single-molecule sequencing assay for the comprehensive profiling of T4 DNA ligase fidelity and bias during DNA end-joining
|
Publications from other scientists
Below is a selection of the latest external publications utilizing NEB’s ligase fidelity data and tools.
|
FRAGLER: A Fragment Recycler Application Enabling Rapid and Scalable Modular DNA Assembly Oling, D., et al. (2022) Describes a comprehensive and robust DNA assembly framework to support rapid and cost-efficient construct generation at scale. The modular system combines features of computationally optimized junctions (NEB Data-optimized Assembly Design), Gly/Ser/Ala codon-junctions (EMMA, MoPET) and a novel software application named FRAGLER (FRAGment recycLER) to perform a variety of functions. |
|
|
Volke, D. C., et al. (2022) Developed a widely-applicable, and high-efficiency genome engineering toolset for Gram-negative bacteria. Tailored a CRISPR base editor that enables single-nucleotide resolution manipulations. Incorporated Cas6-mediated processing of guide RNAs in a streamlined protocol for plasmid assembly using Golden Gate Assembly and NEB Data-optimized Assembly Design, supporting multiplex base editing. |
|
|
PARA: A New Platform for the Rapid Assembly of gRNA Arrays for Multiplexed CRISPR Technologies Yuan, G., et al. (2022) Developed a novel method, PARA, which allows for the one-step Golden Gate Assembly of multiple guide RNAs (gRNAs) into a CRISPR vector with up to 18 gRNAs. Utilized NEB Data-optimized Assembly Design and ligase fidelity data. |
|
|
GEM-Gate: A Low-Cost, Flexible Approach to BioBrick Assembly Bower, C., et al. (2023) Presents a method for synthetic biologists to convert BioBrick parts into parts for Golden Gate Assembly utilizing NEB Data-optimized Assembly Design. |
|
|
Cierniak, F., et al. (2022) Developed a modular chimeric reporter replicon system based on cell culture-adapted and rabbit-derived Zoonotic hepatitis E virus (HEV) strains. Utilized NEB ligase fidelity data for selection of optimal overhangs in high-efficiency Golden Gate Assembly. |
|
|
Stuttmann, J., et al. (2021) Developed a toolkit, based on a highly intron-optimized zCas9i gene, which allows assembly of nuclease constructs expressing up to 32 single guide RNAs (sgRNAs). The toolkit was used to explore the limits of multiplexing in two major model species and provides perspective on how multiplexing can be used to generate complex genotypes or to functionally interrogate groups of candidate genes. Utilized NEB Ligase Fidelity tools for Golden Gate Assembly. |
|
|
Efficient golden gate assembly of DNA constructs for single molecule force spectroscopy and imaging Bell, N. A. W. and J. E. Molloy (2022) Developed a simple and fast technique for making a diverse range of designed DNA structure constructs by combining PCR amplicons and synthetic oligonucleotides using Golden Gate Assembly rules. Demonstrated high yield fabrication of torsionally-constrained duplex DNA up to 10 kb in length and a variety of DNA hairpin structures. |
|
|
Szent-Gyorgyi, C., et al. (2022) Presents the bottom-up design and plasmid synthesis to prepare 10 kb functional yeast secretion and display plasmids that uses an optimized version of Golden Gate Assembly in combination with fluorogen-activating protein reporter technology. Helps address challenging cloning strategies of single plasmids that rely on combinatorial co-expression of a multitude of target and bait fusion reporters useful in projects like library screens. Utilizes NEB Data-optimized Assembly Design principles. |
|
|
Philipp, N., et al. (2023) Introduces DIGGER-Bac, a toolbox for Design and Identification of seed regions for Golden Gate assembly and Expression of synthetic sRNAs in Bacteria. The toolbox supports the process of seed region/scaffold identification (SEEDling) and guides the primer design for high-fidelity Golden Gate cloning (G-GArden). Utilizes NEB Data-optimized Assembly Design principles and Ligase Fidelity Tools. |
|
|
Novel High-Throughput DNA Part Characterization Technique for Synthetic Biology Bak, S. K., et al. (2022) Presents a novel DNA part characterization technique that increases throughput by combinatorial DNA part assembly, solid plate-based quantitative fluorescence assay for phenotyping, and barcode tagging-based long-read sequencing for genotyping. Using the techniques, forty-four DNA parts (21 promoters and 23 RBSs) were successfully characterized in 72 h without any automated equipment. |
|
|
Malci, K., et al. (2022) A review of the molecular methods and SynBio toolkits developed for S. cerevisiae and other emerging nonconventional yeast species. Includes Golden Gate Assembly and use of NEB Ligase Fidelity Tools. |
|
|
Damalas, S. G., et al. (2020) Introduced the SEVA 3.1 platform consisting of the SEVA 3.1 vectors and the Golden Gate-based ‘SevaBrick Assembly’. This platform enables the convergence of standard processes between the SEVA platform, the BioBricks and the Type IIS-mediated DNA assemblies to reduce complexity and optimize compatibility between parts and methods. Utilized NEB ligase fidelity data for selection of optimal overhangs in high-efficiency Golden Gate Assembly. |
|
|
AssemblyTron: flexible automation of DNA assembly with Opentrons OT-2 lab robots Bryant, J. A., Jr., et al. (2023) Presents AssemblyTron, an open-source Python package to integrate j5 DNA assembly design software outputs with build implementation in Opentrons liquid handling robotics with minimal human intervention. Demonstrated the versatility of AssemblyTron through several scarless, multipart DNA assemblies, beginning from fragment amplification, and including Golden Gate Assembly. |
|
|
Polysaccharide II Surface Anchoring, the Achilles' Heel of Clostridioides difficile Malet-Villemagne, J., et al. (2023) Provides new tools to reveal the role of essential genes in C. difficile and finds potential new targets to combat C. difficile infection. Utilized NEB Golden Gate Assembly techniques and tools. |
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/ trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Try our free online tools for Golden Gate Assembly
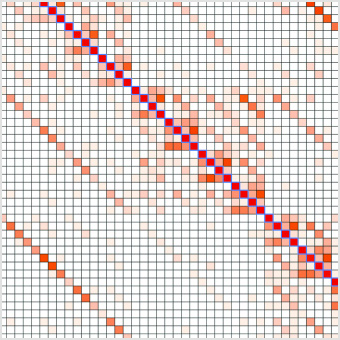
NEBridge Ligase Fidelity ToolsVisualize overhang ligation preferences, predict high fidelity junction sets and split DNA sequences for scarless high-fidelity assembly. |
|
| Try Our Tool |
|
NEBridge Golden Gate
|
|
| Try Our Tool |
|
 Ligase fidelity data & tools: Enabling new applications in synthetic biology
Ligase fidelity data & tools: Enabling new applications in synthetic biology



